Projects
Some of the projects I have parcicipated in, both laboratory research and coursework included.

Single-cell chromatin accessibility analysis reveals the epigenetic basis and signature transcription factors for the molecular subtypes of colorectal cancers
Advised by Prof. Fuchou Tang and Dr. Xin Zhou; collaborate with Dr.Yuqiong Hu, Dr. Haoling Xie, and Dr. Kexuan Chen.
Single-cell chromatin accessibility analysis reveals the epigenetic basis and signature transcription factors for the molecular subtypes of colorectal cancers

Colorectal cancer (CRC) is a highly heterogeneous disease, with well-characterized subtypes based on genome, DNA methylome, and transcriptome signatures. To chart the epigenetic landscape of CRCs, we generated a high-quality single-cell chromatin accessibility atlas of epithelial cells for 29 patients. Abnormal chromatin states acquired in adenomas were largely retained in CRCs, which were tightly accompanied by opposite changes of DNA methylation. Unsupervised analysis on malignant cells revealed two epigenetic subtypes, exactly matching iCMS classification, and key iCMS-specific transcription factors were identified, including HNF4A, PPARA for iCMS2 tumors, and FOXA3, MAFK for iCMS3 tumors. Notably, subtype-specific TFs bind to distinct target gene sets and contribute to both inter-patient similarities and diversities for both chromatin accessibilities and RNA expressions. Moreover, we identified CpG-island methylator phenotypes and pinpointed chromatin state signatures and TF regulators for CIMP-High subtype. Our work systematically revealed the epigenetic basis of the well-known iCMS and CIMP classifications of CRCs.
Source code available at https://github.com/liuzhenyu-yyy/CRC_Epi_scATAC.
For more information, please check our publication at Cancer Discovery.
Read less
High-throughput and high-sensitivity full-length single-cell RNA-seq analysis on third-generation sequencing platform
Advised by Dr. Fuchou Tang; collaborate with Yuhan Liao, Yu Zhang.
High-throughput and high-sensitivity full-length single-cell RNA-seq analysis on third-generation sequencing platform

The advancement of single-cell RNA-seq technologies based on third-generation sequencing (TGS) platforms has accelerated biological researches. Several TGS platform-based single-cell RNA-seq methods have been developed since 2016. Limited by low accuracy and sensitivity, they either combine NGS platform-based methods to reduce the error rates, or sacrifice the throughput to improve detection rates. To overcome these drawbacks, here we developed SCAN-seq2, a high-throughput, high-sensitivity full-length single-cell RNA-seq method based on TGS platform. We performed SCAN-Seq2 to a total of 5,472 cells from nine cell lines. Detailed flowchart of the data processing pipeline and overall statistics of reads after filtering, demultiplexing and deduplication were shown. Through reference-guided transcriptome assembly, we identified thousands of novel full-length RNA isoforms. Transcripts of pseudogenes could be distinguished from transcripts of corresponding parent genes and hundreds of them showed cell type specific expression patterns. Moreover, we showed that V(D)J rearrangement events could be accurately determined for the highly polymorphic TCR and BCR genes (immunoglobins). Finally, we demonstrated the conserved apoptosis response of HepG2 and Hela cells after treated by spliceosome inhibitor Isoginkgetin (IGG). SCAN-seq2 proves to be a new promising tool for single cell full-length transcriptome research.
Source code available at https://github.com/liuzhenyu-yyy/SCAN-seq2.
For more information, please check our publication at Cell Discovery.
Read less
Single-cell Sequencing Reveals Clearance of Blastula Chromosomal Mosaicism in IVF Babies
Advised by Dr. Fuchou Tang, Dr. Yuanqing Yao; collaborate with Yuan Gao, Shuyue Qi.
Single-cell Sequencing Reveals Clearance of Blastula Chromosomal Mosaicism in IVF Babies

While chromosomal mosaic embryos detected by trophectoderm (TE) biopsy offer healthy embryos available for transfer, high resolution postnatal karyotyping and chromosome testing of the transferred embryos are insufficient. Here, we applied single-cell multiomics sequencing for seven infants with blastula chromosomal mosaicism detected by TE biopsy. The chromosome ploidy was examined by singlecell genome analysis with the cellular identity being identified by single-cell transcriptome analysis. A total of 1,616 peripheral leukocyte from seven infants with embryonic chromosomal mosaicism and three control ones with euploid TE biopsy were analyzed. A small number of blood cells showed copy number alterations (CNAs) on seemingly random locations at a frequency of 0-2.5% per infant. However, none of the cells showed CNAs that were with the same as those of the corresponding TE biopsies. The blastula chromosomal mosaicism may be fully self-corrected probably through selective losing of the aneuploid cells during development and the transferred embryos can be born as euploid infants without mosaic CNAs corresponding to the TE biopsies. The results provide a new reference for the evaluations of transferring chromosomal mosaic embryos at certain situations.
Source code available at https://github.com/liuzhenyu-yyy/IVF_Mosaic_Embryo_Transfer.
For more information, please check our publication at Genomics, Proteomics & Bioinformatics.
Read less
Sequential and Distinct Roles of Ngn3 and Neurod1 in Pancreatic Lineage Differentiation
Advised by Dr. Chengran Xu; collaborate with Liu Yang.
Sequential and Distinct Roles of Ngn3 and Neurod1 in Pancreatic Lineage Differentiation

Cell identities are established through the sequential action of key transcription factors (TFs). The current model of pancreas endocrine lineage differentiation invokes the transient expression of basic helix-loop-helix (bHLH) transcription factor Neurogenin 3 (Ngn3) in the trunk cells as the trigger of endocrine fate commitment and rapid differentiation to pro-islet cells. Another bHLH transcription factor named Neurod1, which is directly regulated by Ngn3, also plays critical roles in the pancreatic endocrine specification. Despite the key regulatory function of these two TFs, their genomic targets and the regulatory mechanism remains largely unknown. Here, we had generated a new transgenic mouse strain to uncover the genome-wide direct target of Ngn3 with the CUT&RUN method and performed comprehensive investigations of the sequential and distinct chromatin regulatory program mediated by Ngn3 and Neurod1. We found that Ngn3 access genome enhancers in highly condensed chromatin regions and prompt their activation. The enhancer landscape established by Ngn3 is further maintained by the expression of Neurod1. Neurod1 also binds the promoter region of key endocrine TFs to help initiate their expression. These data suggest the pioneering ability of Ngn3, with Neurod1 acting as a co-factor to maintain the opened chromatin state.
Read less
Chaotic Model for Insulin-Glucose System
A chaotic model for the insulin-glucose regulatory system. Using this model, we could mimic several common metabolic disorders related to the system and identify key parameters responsible for the initiation of these disease.
Description
This is my final project in “Mathematical Modeling in the Life Sciences” Course of Prof. Louis Tao in the spring semaster of 2020. Here a novel mathematical model based on Lotka–Volterra equations was developed to mimic the behaviors of the insulin-glucose system. Numerical simulation indicates that the model has various different dynamic behaviors under different conditions, including chaos. Using this model, we could mimic several common metabolic disorders related to the system and identify key parameters responsible for the initiation of these disease.
Model presentation
Alfred Lotka and Vito Volterra proposed the two-dimension ordinary differential equation model to describe the population dynamics of prey and predator in 1926. Prompted by their model, the relationship of glucose and insulin is also like prey and predator. It’s reasonable to model the insulin-glucose system by making some adjustments to the Lotka–Volterra model.
The mathematical relationships for the model are formulated as follows:

Where x(t) is the concentration of insulin, y(t) is the population of glucose and z(t) is the population density of β-cells. There are 21 distinct model parameters, each has specific biological meaning, as discussed in project report, and could be adjusted to simulate both normal condition and metabolic disorders.
More information about the model, including time series and state space, stability analysis, bifurcation diagrams and chaotic analysis were avaliable at the project page.
Read less
Game of the Amazons AI
An SVM-based classifier to predict genome enhancers based on ChIP-Seq and ATAC-Seq signals.

Description
This is my final project in “Methods in Bioinformatics” Course of Prof. Ge Gao in the autumn semaster of 2019. Here I designed a novel classifier to predict genome enhancers. The imput features include chromatin modification and related enzymes, transcription factor binding and chromatin accessibility. Features were selected using random forest and classifier was designed with supporting vector machine (SVM). A 10-fold cross validation was performed for parameter tuning under several different regression methods and sigmoid functions. The final model acquires a precision of 0.9886 and a recall of 0.9931 on K562 cell lines, with the $F_1$ score being 0.9908.
Command Line
Trained model was saved in RData format and could be used with a single integrated script EnhancerPredictor.R. The most simple command to run Enhancer Predictor:
Rscript EnhancerPredictor.R input.bed output.bed
Check more at the project page.
Read less
Game of the Amazons AI
A simple DFS-based bot for the Game of the Amazons, with optimized scoring function.
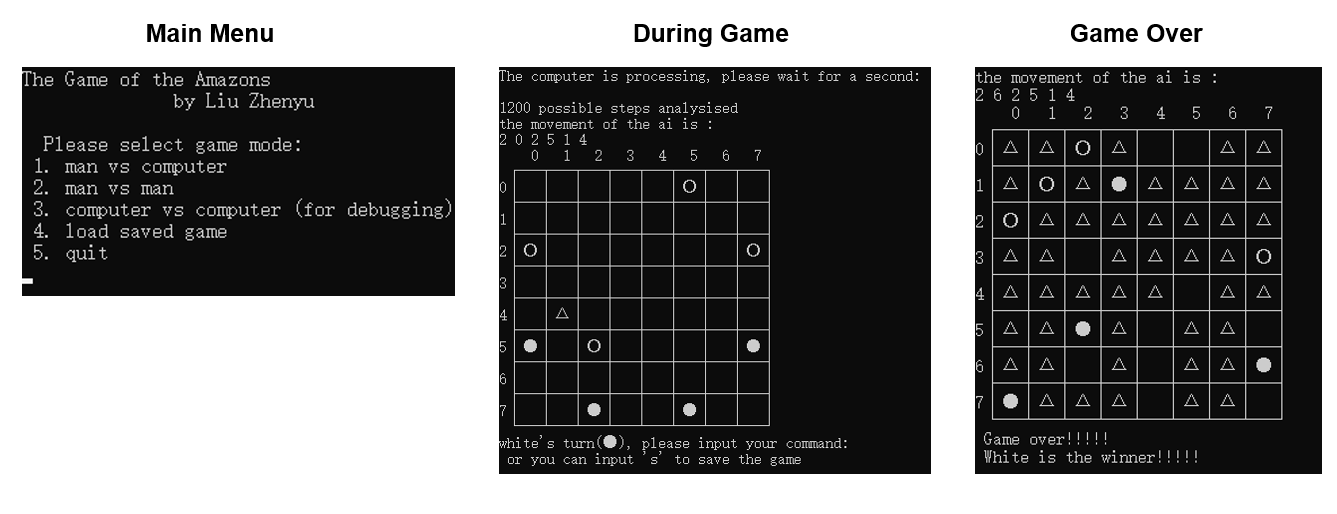
Description
This is my code for the Amazons Lab in “Introduction to Computation (A)” Course of Prof. Yafei Dai in the autumn semester of 2018. The Amazons Lab required us to develop a c/c++ program for the Game of the Amazons with following features:
- Main menu for selection (new game, save game, start new game, quit);
- Chess board and piece indicated by ASCII art;
- Allowing player versus player and player versus computer;
- Able to save current game and load saved games.
The software utilze depth-first search to identify the best movement at current situation, as achieved by recursive and recursion of several different chess moving functions. I only designed single-layed search and performs no pruning step during searching, which could be further improved.
Check more at the project page.
Read less
Tang Lab Quota Usage
A shiny app to visualize quota usage for each user from Tang Lab on PKUHPC.
Tang Lab Quota Usage

A shiny app to visualize quota usage for each user from Tang Lab on PKUHPC.
URL: https://liuzhenyu-yyy.shinyapps.io/tanglab_quota/
Source code available at https://github.com/liuzhenyu-yyy/TangLab-Quota.
Web appwas deployed on shinyapps.io.
Read less